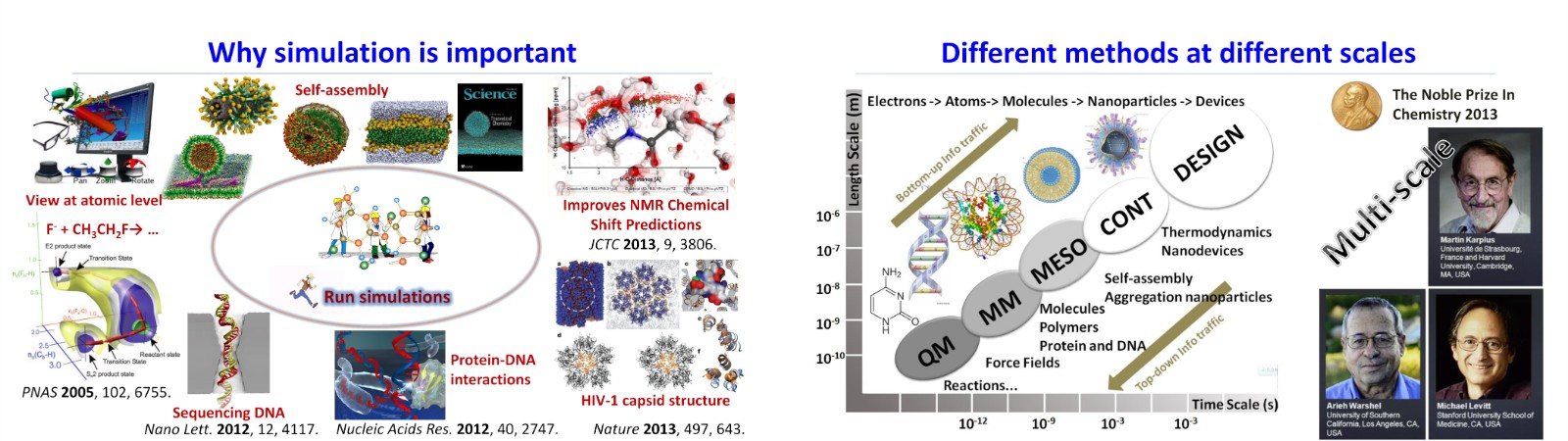
Theoretical calculations and multi-scale simulations are powerful tools in understanding biological processes and the properties of soft matter systems at the atomistic level. We uses various methods ranging from ab initio (DFT) simulations to mesoscopic simulations, including atomistic and coarse-grained molecular dynamics, to investigate the self-assembly of spherical nucleic acids (SNAs) driven with DNA-based nanomachines and to understand the curvature-mediated interactions. These methods are also used to explore the physical properties of nucleic acids and the biophysics of toehold-mediated strand displacement reaction. We also collaborate with experimentalists to study red‐light‐mediated photoredox catalysis and the interactions of polymer moleculecules with membranes. Our current research projects are listed as following.
(1) The biophysics of toehold-mediated strand displacement reaction and DNA reaction network.
(2) The nanoparticle (NP) assembly directed using DNA molecular machines.
(3) Membrane-mediated interactions.
(4) The selective interactions of polymer molecules with membranes.
多尺度理论模拟可以为理解和研究生物过程、软物质结构和特性等提供原子水平的研究视角。我们长期关注理论模拟方法的发展,及其在高分子物理化学、软物质物理、以及生物物理中的应用,研究手段包括但不限制于从第一性原理到介观尺度分子模拟,包括全原子和粗粒化分子动力学模拟等。
近年来,我们的研究兴趣主要集中在软物质结构跨尺度调控等方面。包括:DNA反应网络调控的球形核酸组装,膜曲率介导相互作用、大分子的结构和功能等。具体的课题包括:(1) 链替换反应微观物理机制;(2)DNA反应网络及纳米粒子组装;(3)膜曲率介导相互作用;(4)生物膜与纳米粒子相互作用等。
一直以来,我们十分关注方法和技术的发展,努力引入和发展一些新的模拟方法和理论模型,包括Energy landscape等;同时,针对感兴趣的研究方向和课题,我们一直与实验(实验课题组)保持紧密的合作。
56. G. Chen, X. Li, Y. Huang, C. Zheng, H. Fang, Q. Zhang, K. Liu, K. Wu, Q. Zhu, C. Guo, H. Zhu, R. Dai, Y. He, Q. Zhu, W. Zhou, Y. Yang, Z. Li, K. Qu*, B. Li*, S. Xiao*, S. Tian, B. Xing, X. Chen, A. Z. Wang, Y. Xiong, Yuanzeng Min*; Platinonitrene-based radiosensitizer boosts radiotherapy and abscopal effect, Nat. Biomed. Eng, 2025, Accepted.
55. Z. Li, C. Li, S. Xiao*, H. Liang; Efficient and Precise Integration of Large DNA Sequences Using Precise Interstrand Cross-Linking of Long ssDNA and sgRNA; ACS Synth. Biol 2025, 14(5), 1451–1463.
54. S. Zhou#, P. Lyu#, Y. Huang, X. Man*, S. Xiao*; Nonmonotonous Translocation Dynamics of Highly Deformable Particles across Channels; ACS Nano 2025, 19, 10807–10815.
53. Y Liang#, Y Zhang#, Y Huang#, C Xu#, J Chen, X Zhang, B Huang, Z Gan, X Dong, S Huang, C Li, S Jia, P Zhang, Y Yuan, H Zhang, Y Wang, B Yuan, Y Bao*, S Xiao*, M Xiong*; Helicity-directed recognition of bacterial phospholipid via radially amphiphilic antimicrobial peptides; Sci. Adv. 2024, 10, eadn9435.
52. Z Li, Y Ma, C Li, S Xiao*, H Liang; Photo‐Cross‐linked DNA Structures Greatly Improves Their Serum Nuclease Resistances and Gene Knock‐In Efficiencies. Small Methods. 2024, 2401346. (邀稿,Wiley MaterialsViews 官方报道: https://mp.weixin.qq.com/s/ti3xtnDOZeeRvzvHPOqdOw)
51. C Li, S Xiao*, H Liang*; The effects of fluorescent pair on the kinetics of DNA strand displacement reaction; Chinese J. Chem. Phys. 2024, 37 (5), 679-684.
50. Y Zhang, S Xiao*, H Liang*; Avoiding Kinetic Trapping in the Self-Assembly of DNA-Functionalized Gold Nanoparticles by Using an Enthalpy-Mediated Strategy; Macromolecules 2023, 56 (9), 3454-3463.
49. X Yang, W Shi, Z Chen, M Du, S Xiao*, S Qu*, C Li*; Light-Fueled Nonequilibrium and Adaptable Hydrogels for Highly Tunable Autonomous Self-Oscillating Functions; Adv. Funct. Mater. 2023, 33 (24), 2214394.
48. Y Du, Y Zhang, J. Jin*, S. Xiao*, H Liang, W Jiang*; Topology-Directed Self-Locking of Colloidal Suprastructures; Macromolecules 2023, 56 (9), 2781–2789.
47. S Xiao*#, C. Li#, H Liang*; DNA: structure, strand displacement and reaction network; SCIENTIA SINICA Chimica 2023, doi: 10.1360/SSC-2023-0006. (邀稿)
46.T Chen, Y Zhang*, S Xiao, D Yao, W Jiang*, H Liang*; Catalytic assembly of symmetric diblock copolymers in a thin film: A dissipative particle dynamics simulation; Phys. Chem. Chem. Phys. 2023, 25 (14), 9779-9784.
45. M. Liu#, L. Huang#, W. Zhang#, X. Wang, Y. Geng, Y. Zhang, L. Wang, W. Zhang, Y. Zhang, S. Xiao*, Y. Bao*, M. Xiong*, and J. Wang*; A transistor-like pH-sensitive nanodetergent for selective cancer therapy; Nat. Nanotechnol. 2022, 17 (5), 541-551.
44. Z Chen, S Zheng, Z Shen, J Cheng, S Xiao*, G Zhang, S Liu*, J Hu*; Oxygen-Tolerant Photoredox Catalysis Triggers Nitric Oxide Release for Antibacterial Applications; Angew. Chem. Int. Ed. 2022, 61 (30), e202204526.
43. C. Li, Z. Li, W. Han, X. Yin, X. Liu, S. Xiao*, and H. Liang*; How the Fluorescent Labels Affect the Kinetics of Toehold-Mediated DNA Strand Displacement Reaction?; Chem. Commun. 2022, 58, 5849-5852.
42. C Li, S Xiao*, H. Liang*; Regulation of Gene Expression via Toehold Switch in E. coli; J. Funct. Polym. 2023, 36(3), 302-309.
41. H Zhang, S Shan, Y Huang, S Xiao, D Chen, G Zou*; Regulable chiral amplification effects in copoly (phenylacetylene) s and bidirectional manipulation for helix preferences; J. Mater. Chem. C 2022, 10 (38), 14265-14272.
40. Z. Shen, S. Zheng, S. Xiao*, R. Shen, S. Liu*, and J. Hu*; Red-Light-Mediated Photoredox Catalysis Enables Self-Reporting Nitric Oxide Release for Efficient Antibacterial Treatment; Angew. Chem. Int. Ed. 2021, 60 (37), 20452-20460.
39. T. Chen, Y. Zhang, X. Li, C. Li, T. Lu, S. Xiao*, and H. Liang*; Curvature-Mediated Pair Interactions of Soft Nanoparticles Adhered to a Cell Membrane; J. Chem. Theory Comput. 2021, 17 (12), 7850-7861.
38. J. Li, Z. Ge*, K. Toh, X. Liu, A. Dirisala, W. Ke, P. Wen, H. Zhou, Z. Wang, S. Xiao, J. F. R. Van Guyse, T. A. Tockary, J. Xie, D. Gonzalez-Carter, H. Kinoh, S. Uchida, Y. Anraku, and K. Kataoka*; Enzymatically Transformable Polymersome-Based Nanotherapeutics to Eliminate Minimal Relapsable Cancer; Adv. Mater. 2021, 33 (49), 2105254.
37. Y. Guo, D. Yao*, B. Zheng, X. Sun, X. Zhou, B. Wei, S. Xiao, M. He, C. Li, and H. Liang*; pH-Controlled Detachable DNA Circuitry and Its Application in Resettable Self-Assembly of Spherical Nucleic Acids, ACS Nano 2020, 14, 8317–8327.
36. B. Wei, X. Sun*, D. Yao, C. Li, S. Xiao, Y. Guo, and H. Liang*; Homogeneous DNA-only keypad locks enable one-pot assay of multi-inputs,Chem. Commun. 2020, 56, 7427-7430.
35. X Zhou#, D. Yao#, W. Hua, N. Huang, X. Chen, L. Li*, M. He, Y. Zhang, Y Guo, S. Xiao*, F., Bian*, and H. Liang*; Programming colloidal bonding using DNA strand-displacement circuitry, PNAS 2020, 117 (11), 5617-5623.
34. S Xiao, D Sharpe, D Chakraborty, D Wales*; Energy Landscapes and Hybridization Pathways for DNA Hexamer Duplexes; J. Phys. Chem. Lett. 2019, 10 (21), 6771-6779.
33. S Xiao, H Liang*, D Wales*; The Contribution of Backbone Electrostatic Repulsion to DNA Mechanical Properties is Length-Scale-Dependent; J. Phys. Chem. Lett. 2019, 10, 4829-4835.
32. C He#, M Wang#, X Sun, Y Zhu, X Zhou, S Xiao*, Q Zhang, F Liu, Y Yu, H Liang, G Zou*; Integrating PDA microtube waveguide system with heterogeneous CHA amplification strategy towards superior sensitive detection of miRNA; Biosens. Bioelectron. 2019, 129, 50-57.
31. T Zuo, C Ma, G Jiao, Z Han, S Xiao, H Liang, L Hong, D Bowron*, A Soper, C C. Han, H Cheng*; Water/Cosolvent Attraction Induced Phase Separation: A Molecular Picture of Cononsolvency; Macromolecules 2019, 52 (2), 457-464.
30. H Tang, Z Gu, H Ding, Z. Li, S Xiao, W. Wu, X Jiang*; Nanoscale Crystalline Sheets and Vesicles Assembled from Non-Planar Cyclic pi-Conjugated Molecules; Research 2019, 1953926.
29. B Wei, D Yao*, B Zheng, X Zhou, Y Guo, X Li, C Li, S Xiao, H Liang*; A Facile Strategy for Visible Disassembly of Spherical Nucleic Acids Programmed by Catalytic DNA Circuit; ACS Appl. Mater. Interfaces 2019, 11 (22), 19724-19733.
28. X Li, X Sun, J Zhou, D Yao, S Xiao, X Zhou, B Wei, C Li, H Liang*; Enzyme-assisted waste-to-reactant transformation to engineer renewable DNA circuits; Chem. Commun. 2019, 55 (77), 11615-11618.
27. X Sun, B Wei, Y Guo, S Xiao*, X Li, D Yao, X Yin, S Liu, H Liang*; A Scalable “Junction Substrate” to Engineer Robust DNA Circuits; J. Am. Chem. Soc. 2018, 140 (31), 9979–9985. (被选为当期JACS封面文章)
26. L Chen, X Li, Y Zhang, T Chen, S Xiao* and HJ Liang*; Morphological and mechanical determinants of cellular uptake of deformable nanoparticles; Nanoscale 2018, 10, 11969-11979.
25. Y Zhang, T Chen, Z Pan, X Sun, X Yin, M He, S Xiao*, H Liang; Theoretical Insights into the Interactions between Star-shaped Antimicrobial Polypeptides and Bacterial Membrane; Langmuir 2018, 34 (44), 13438-13448.
24. X Zhou#, D Yao#, M He, S Xiao*, H Liang*; Optimizing the Toehold Strategy of On-Chip Nucleic Acid Hybridization Probe for the Discrimination of Single Nucleotide Polymorphism; Langmuir 2018, 34 (49), 14811-14816.
23. Y Guo#, B Wei#, X Sun, D Yao, X Zhou, S Xiao*, H Liang*; DNA and DNA computation based on toehold-mediated strand displacement reactions; Int. J. Mod. Phys. B 2018, 32, 1840014.
22. X Zhou#, H Li#, M He, X Yin, D Yao, S Xiao, H Liang*; Photoresponsive spherical nucleic acid: spatiotemporal control of the assembly circuit and intracellular microRNA release; Chem. Commun. 2018, 54, 106-109.
21. J Li#, S Xiao#, Y Xu, S Zuo, Z Zha, W Ke, C He*, Z-S Ge*; Smart Asymmetric Vesicles with Triggered Availability of Inner Cell-Penetrating Shells for Specific Intracellular Drug Delivery; ACS Appl. Mater. Interfaces 2017, 9 (21), 17727–17735. (# 共同一作)
20. D Yao, S Xiao*, X Zhou, H Li, B Wang, B Wei and H Liang*; Stacking Modular DNA Circuitry in Cascading Self-Assembly of Spherical Nucleic Acids; J. Mater. Chem. B 2017, 5, 6256–6265. (“Hot Paper” theme of J. Mater. Chem. B)
19. Y Guo, B Wei, S Xiao, D Yao, H Li, H Xu, T Song, X Li, H Liang*; Recent advances in molecular machines based on toehold-mediated strand displacement reaction; Quantitative Biology 2017, 5 (1), 25–41.
18. L Chen, S Xiao*, H Zhu, L Wang and H Liang; Shape-dependent internalization kinetics of nanoparticles by membranes; Soft Matter 2016, 12 (9), 2632-2641.
17. D Yao, H Li, Y Guo, X Zhou, S Xiao* and H Liang*; A pH-responsive DNA nanomachine controlled catalytic assembly of gold nanoparticles; Chem. Commun. 2016, 52, 7556-7559.
16. S Xiao and H Liang*; DNA and DNA computation based on toehold-mediated strand displacement reactions; Acta Phys. Sin. 2016, 65 (17), 178106.
15. D Yao, T Song, X Sun, S Xiao*, F Huang and H Liang*; Integrating DNA Strand Displacement Circuitry with Self-Assembly of Spherical Nucleic Acids; J. Am. Chem. Soc. 2015, 137 (44), 14107-14113.
14. D Yao, B Wang, S Xiao*, T Song, F Huang and H Liang*; What Controls the “Off/On Switch” in the Toehold-Mediated Strand Displacement Reaction on DNA Conjugated Gold Nanoparticles? Langmuir 2015, 31 (25), 7055–7061.
13. D Yao, T Song, B Zheng*, S Xiao, F Huang, H Liang*; The combination of gold nanorods and nanoparticles with DNA nanodevices for logic gates construction; Nanotechnology 2015, 26 (42), 425601.
12. H Li, S Xiao, D Yao, M H W Lam* and H Liang* ; A Smart DNA-Gold Nanoparticle Probe for Detecting Single-base Changes on the Platform of Quartz Crystal Microbalance; Chem. Commun. 2015, 51, 4670-4673.
11. T Song, S Xiao*, F Huang, M Hu, and H Liang*; An efficient DNA-fueled molecular machine for the discrimination of single-base changes; Adv. Mater. 2014, 26 (35), 6181-6185.
10. S Xiao, H Zhu, L Wang, L Chen and H Liang*; Enhancing the efficiency of lithium intercalation in carbon nanotube bundles using surface functional groups; Phys. Chem. Chem. Phys. 2014, 16, 16003-16012.
9. S Xiao, H Zhu, L Wang, and H Liang*; DNA conformational flexibility study using phosphate backbone neutralization model; Soft Matter 2014, 10, 1045-1055.
8. S Xiao, M. L. Klein, D. N. Lebard, B. G. Levine, H Liang, M. Alfonso-Prieto*; Magnesium-dependent RNA Binding to the PA Endonuclease Domain of the Avian Influenza Polymerase; J. Phys. Chem. B 2014, 118, 873-889.
7. F Huang, X Zhou, D Yao, S Xiao and H Liang*; DNA Polymer Brush Patterning through Photocontrollable Surface‐Initiated DNA Hybridization Chain Reaction; Small 2015, 11 (43), 5800-5806.
6. H Zhu, S Xiao, L Wang, and H Liang*; Asymmetrical Cation Movements through G-quadruplexes DNA; J. Chem. Phys. (Communication) 2014, 141, 041103.
5. H Xu, W Deng, F Huang*, S Xiao, G Liu, H Liang*; Enhanced DNA toehold exchange reaction on a chip surface to discriminate single-base changes; Chem. Commun. 2014, 50 (91), 14171-14174.
4. L Wang, N Li, S Xiao, and H Liang*; Thermodynamic and conformational insights into the phase transition of a single flexible homopolymer chain using replica exchange Monte Carlo method; J. Mol. Modeling 2014, 20, 1-10.
3. H Zhu, S Xiao and H Liang*; Structural Dynamics of Human Telomeric G-quadruplex Loops Studied by Molecular Dynamics Simulations; PLoS ONE 2013, 8 (8), e71380.
2. S Xiao and H Liang*; The conformational flexibility of nucleic acid bases paired in gas phase: A Car-Parrinello molecular dynamics study; J. Chem. Phys. 2012, 136, 205102. (selected for the June 1, 2012 issue of Virtual Journal of Biological Physics Research.)
1. S Xiao, L Wang, Y Liu, X Lin and H Liang*; Theoretical investigation of the proton transfer mechanism in guanine-cytosine and adenine-thymine base pairs; J. Chem. Phys. 2012, 137, 195101.
* Google scholar citations: http://scholar.google.com/citations?user=Jo0QvogAAAAJ
- 梁好均 ,姚东宝,肖石燕,宋廷结,一种DNA催化循环网络诱导的DNA-纳米金组装方法及其在单碱基突变检测中的应用,201510350472.4,2015/10/28,2017/08/29.
- 梁好均 ,宋廷结,肖石燕,一种DNA分子机器和基于DNA分子机器的单碱基突变检测方法及其应用,201310471003.9,2014/02/12,
- 暂无内容
