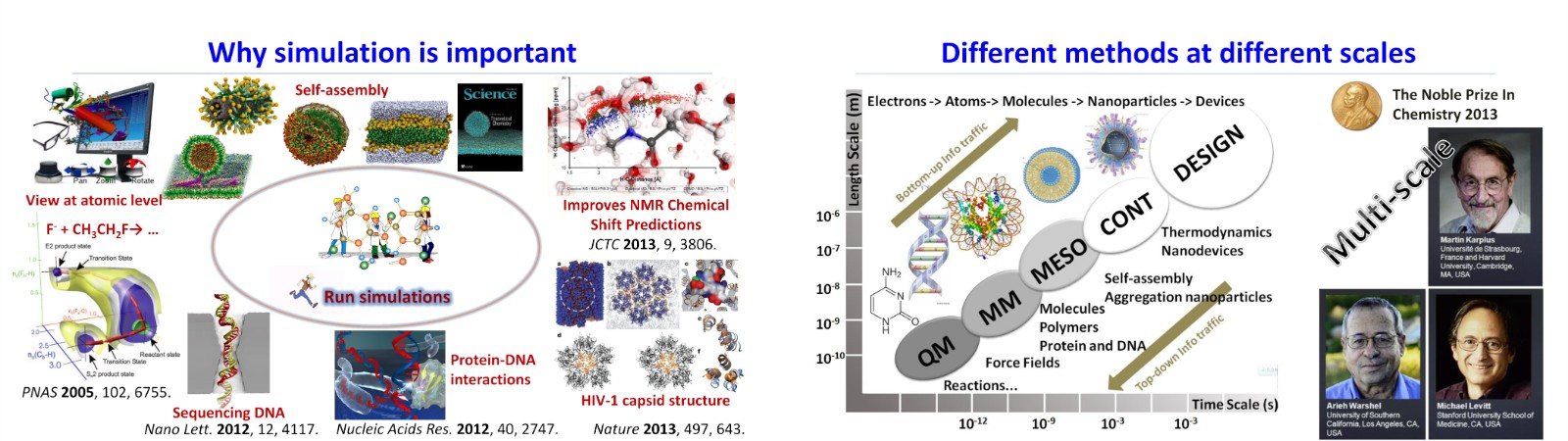
Theoretical calculations and multi-scale simulations are powerful tools in understanding biological processes and the properties of soft matter systems at the atomistic level. We uses various methods ranging from ab initio (DFT) simulations to mesoscopic simulations, including atomistic and coarse-grained molecular dynamics, to investigate the self-assembly of spherical nucleic acids (SNAs) driven with DNA-based nanomachines and to understand the curvature-mediated interactions. These methods are also used to explore the physical properties of nucleic acids and the biophysics of toehold-mediated strand displacement reaction. We also collaborate with experimentalists to study red‐light‐mediated photoredox catalysis and the interactions of polymer moleculecules with membranes. Our current research projects are listed as following.
(1) The biophysics of toehold-mediated strand displacement reaction and DNA reaction network.
(2) The nanoparticle (NP) assembly directed using DNA molecular machines.
(3) Membrane-mediated interactions.
(4) The selective interactions of polymer molecules with membranes.
多尺度理论模拟可以为理解和研究生物过程、软物质结构和特性等提供原子水平的研究视角。我们长期关注理论模拟方法的发展,及其在高分子物理化学、软物质物理、以及生物物理中的应用,研究手段包括但不限制于从第一性原理到介观尺度分子模拟,包括全原子和粗粒化分子动力学模拟等。
近年来,我们的研究兴趣主要集中在软物质结构跨尺度调控等方面。包括:DNA反应网络调控的球形核酸组装,膜曲率介导相互作用、大分子的结构和功能等。具体的课题包括:(1) 链替换反应微观物理机制;(2)DNA反应网络及纳米粒子组装;(3)膜曲率介导相互作用;(4)生物膜与纳米粒子相互作用等。
一直以来,我们十分关注方法和技术的发展,努力引入和发展一些新的模拟方法和理论模型,包括Energy landscape等;同时,针对感兴趣的研究方向和课题,我们一直与实验(实验课题组)保持紧密的合作。
